TL;DR
Awk crunches massive data; a High Performance Computing (HPC) script calls hundreds of Awk concurrently. Fast and scalable in-memory solution on a fat machine.
Introduction
Presenting the solution I worked on in 2018, to a Data Challenge organized at work. I solve the Scientific Publications Mining challenge (no.4) that consists of 5 problems. I use classic Unix tools with a modern scalable HPC scripting tool to work out the solutions. The project is hosted on github. About 12 teams entered the contest.
Tools
Software
Awk (gawk v4.0.2) is dominantly used for the bulk of core processing.
Argonne National Laboratory developed HPC scripting tool called Swift (NOT the Apple Swift) is used to run the Awk programs concurrently over the dataset to radically improve performance. Swift uses MPI based communication to parallelize and synchronize independent tasks.
Other Unix tools such as sort, grep, tr, sed and bash are used as well. Additionally, jq, D3, dot/graphviz, and ffmpeg are used.
Hardware
Fortunately, I had access to a large-memory (24 T) SGI system with 512-core Intel Xeon (2.5GHz) CPUs. All the IO is memory (/dev/shm) bound ie. the data is read from and written to /dev/shm.
Rationale
Awk is lightweight, concise, expressive, and fast – especially for text processing applications. Some people find Awk programs terse and hard to read. I have taken care to make the code readable. I wanted to see how far can I go with Awk (and boy did I go far!). Alternative tools such as modern Python libraries sometimes have scaling limitations, portability concerns. Some are still evolving. Swift is used simply because I was familiar with it and confident that it will scale well in this case.
Data
The original data was in two sets (aminer
and mag) of 322 json files – each containing a million records. A file
with a list of common records appearing in both sets was available. An Awk script
(src/filterdup.awk) is used to exclude these duplicate records from the aminer
dataset. As a result, it came out about 256 million (256,382,605 to be
exact) unique records to be processed. The total data size is 329GB. Some
fields in the data are null. Those records are avoided where relevant.
Additionally, records related to non-English publications were avoided as
needed. A
snapshot
of tabular data is available. String qwqw is chosen as a column separator to
distinguish it from text already found in data. All other 3 or less character
combinations already existed in data prohibiting them to be used as separators.
#!/usr/bin/env awk -f
# $1 magid
# $2 aminerid
# Filter duplicate papers and remove them
# from aminer database based on the linking relationship
BEGIN {FS = OFS = "qwqw"}
NR == FNR {a[$2] = $1}
!($1 in a) && FILENAME ~ /aminer/ { print }
NR == FNR is a cool Awk idiom that ensures the condition is true only for the first file. This is because for each file that is processed the FNR (File Record Number) gets reset but the NR does not. This means the condition NR == FNR yields true only for the first file.
In addition to the publications data, I use the following:
-
A list of large cities (population 100K+) and their lat-long coordinates (3,517).
-
A list of countries (190).
-
A list of world universities and research institutes (8,984).
-
A list of stop-words to avoid in some of the results (161 words).
Solutions
Pre- and post-processing
jq is used to transform the json data to tabular format
(src/json2tabular.sh). The converted tabular files have 19 original columns
(id, title, authors, year, citations, etc) and one
additional column called num_authors showing the number of authors for a
given publication record. The authors column has a semi-colon separator for
multiple authors. Further curation of tabular data is done by removing
extraneous space, square brackets, escape characters and quotes using sed.
Some of the results obtained were postprocessed for visulization using the D3
graphics framework. ffmpeg is used to stitch images of trending terms to
create an animation. dot/graphviz is used to build the massive citation
network graph of the best paper.
Scaling up
Each solution has Awk code run concurrently over the 322 data files on 322 CPU cores using Swift. This resulted in radical speedup at scale. None of the solution has taken more than an hour of runtime–most took less than a minute.
Problem 1
Identify the individual or group of individuals who appear to be the expert in a particular field or sub-field.
This is solved in two ways. First approach identifies all the entries with
citations higher than 500 for a given search topic
(results/meditation_highly_cited.txt).
#!/usr/bin/env awk -f
BEGIN {
# Field Separator
FS = "qwqw"
# Output field separator
OFS = "\t"
IGNORECASE = 1
# Field names for readability
id=1; title=2; num_authors=3; doi=4; fos_isbn=5; doctype_issn=6;
lang=7; n_citation=8; issue=9; url=10; volume=11; page_start=12;
page_end=13; year=14; venue=15; publisher_pdf=16; references=17;
keywords=18; abstract=19; authors=20;
}
($0~topic && $num_authors > 0 && $n_citation!~/null/ && $n_citation>500){
print $n_citation, $title, $authors, $year
}
# How to run:
# awk -v topic=meditation -f src/prob1_p1.awk data/mag_papers_sample.allcols.txt
The second approach finds the names of authors whose names are repeating for
queried topic with at least a certain number of citations in each entry. This
gives a reasonable idea of who are the expert figures in a given research area.
One such result in results/cancer_research_topauths.txt shows authors in
cancer research with more than one publication with at least 1,000 citations.
#!/usr/bin/env awk -f
BEGIN {
# Field separator
FS = "qwqw"
OFS = "\t"
IGNORECASE = 1
# Field names
id=1; title=2; num_authors=3; doi=4; fos_isbn=5; doctype_issn=6;
lang=7; n_citation=8; issue=9; url=10; volume=11; page_start=12;
page_end=13; year=14; venue=15; publisher_pdf=16; references=17;
keywords=18; abstract=19; authors=20;
}
($0~topic && $num_authors>0 && $n_citation!~/null/ && $n_citation>1000) {
# find the authors whose names are repeating for a particular topic.
# Those authors will be considered experts.
gsub("\"","",$authors)
split($authors, a, ";")
for (i in a) {
split(a[i], b, ",")
# auths array will have keys as auth names and the
# element value increases if the key repeats
if(b[1]!~/null/) auths[b[1]]++
}
}
END { for (k in auths) if(auths[k]>1) print auths[k], k }
#How to run:
# awk -v topic=cancer -f src/prob1_p2.awk data/mag_papers_sample.allcols.txt
# sort the results
The HPC implementation of this solution finishes in 25 seconds.
Alongside is the citation network graph of the most cited paper in this
diagram
(too big to fit here). The result of a query for all-time list of most cited
papers with a threshold of 20,000 is in results/top_papers.txt.
Problem 2
Identify topics that have been researched across all publications.
This is solved by identifying most frequently appearing words in the
collection. Title, abstract and keywords are parsed and top 1,000 frequently
occurring words across the whole collection is found. Several common words (aka
stop-words) are filtered from the results. At over 23 million, the word
“patients” occurs the most frequently. The full list of top 1,000 words is
found in /results/top_1K_words_kw_abs_title.txt. The target collection of
publications may be narrowed down to criteria such as years range.
#!/usr/bin/env awk -f
# Problem Statement
# Identify topics that have been researched across all publications.
# Solution:
# step1. Filter the input to English language records
# step2. Eliminate unnecessary content such as punctuation,
# non-printable chars and small words such as
# 1 letter and 2 letter words
# step3. Extract words used in keywords, title and abstract
# step4. Find most frequently used words
BEGIN {
FS = "qwqw"
IGNORECASE = 1
# Field names
id=1; title=2; num_authors=3; doi=4; fos_isbn=5;
doctype_issn=6;lang=7; n_citation=8; issue=9; url=10;
volume=11; page_start=12; page_end=13; year=14; venue=15;
publisher_pdf=16; references=17; keywords=18; abstract=19; authors=20;
}
#collect stop words
NR == FNR {x[$1];next}
$lang~/en/ && ($keywords!~/null/ || $title!~/null/ || $abstract!~/null/) {
# treat titles
$title = tolower($title)
split($title, a, " ")
for (i in a) if(length(a[i])>2 && match(a[i],/[a-z]/) && a[i] in x == 0) kw[a[i]]++
# treat keywords
$keywords = tolower($keywords)
split($keywords, b, ",")
for (i in b) if(length(b[i])>2 && match(b[i],/[a-z]/) && b[i] in x == 0) kw[b[i]]++
# treat abstracts (Computationally expensive, results are in:
# top_1000_words_from_kw_abstract_title_by_freq.txt)
$abstract = tolower($abstract)
gsub("\"","",$abstract)
gsub(",","",$abstract)
split($abstract, c, " ")
for (i in c) if(length(c[i])>2 && match(c[i],/[a-z]/) && c[i] in x == 0) kw[c[i]]++
}
END {
for(k in kw){
if (kw[k]>1000) print kw[k], k
}
}
# HOW TO RUN: LC_ALL=C awk -f prob2.awk stop_words.txt \
../aminer_papers_allcols_excl/aminer_papers_*.allcols.excl.txt \
../mag_papers_allcols/mag_papers_*.allcols.txt \
| sort -nr > freq.txt
The HPC implementation (Swift code shown below) finishes in 9 minutes.
import files;
import unix;
/* app defines what we want to run, the input parameters,
where the stdout should go, etc.
*/
app (file out) myawk (file awkprog, file stop_words, file infile){
"/usr/bin/awk" "-f" awkprog stop_words infile @stdout=out
}
/* populate the input data */
file aminer[] = glob("/dev/shm/aminer_mag_papers/*.txt");
/* output for each call will be collected here */
file outfiles[];
foreach v, i in aminer {
outfiles[i] = myawk(input("/home/km0/SMC18/src/prob2.awk"),
input("/home/km0/SMC18/data/stop_words.txt"),
v);
}
/* Combine all output in one file */
file joined <"joined.txt"> = cat(outfiles);
/*
After running this swift app:
awk '{a[$2]+=$1} END {for (k in a) print a[k],k}' joined.txt | sort -nr > freq.txt
*/
Problem 3
Visualize the geographic distribution of the topics in the publications.
This is solved by identifying the author affiliations for the records that has the search topic in them. The affiliation is searched against three databases–cities, universities and countries to find out the geographic locations for that research. The results are aggregated to present a list of centers for which a given keyword appears most frequently. For cities, the results are plotted on world map. One such result is shown below for the topic of research on “birds”.
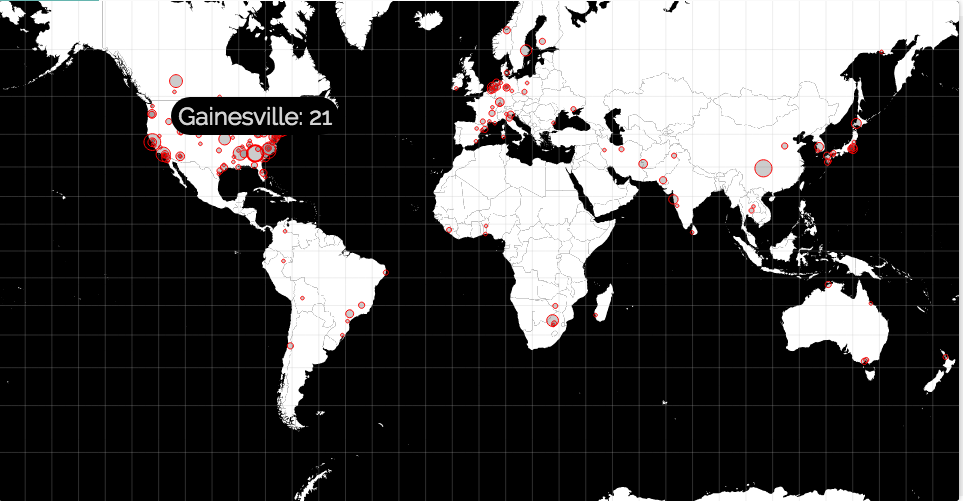
The results/ directory contains other similar results such as epilepsy,
opioid, meditation research by universities and by countries. The HPC
implementation finishes in 25 seconds. The Awk code is shown below.
#!/usr/bin/env awk -f
# problem statement
# visualize the geographic distribution of the topics in the publications.
BEGIN {
# Field separator
FS = OFS = "qwqw"
IGNORECASE = 1
# Field names
id=1; title=2; num_authors=3; doi=4; fos_isbn=5; doctype_issn=6;
lang=7; n_citation=8; issue=9; url=10; volume=11; page_start=12;
page_end=13; year=14; venue=15; publisher_pdf=16; references=17;
keywords=18; abstract=19; authors=20;
}
#collect the countries/cities/univs data
NR == FNR {a[$1];next}
#treat records with authors whose affiliation is available
$0~topic && $num_authors!~/null/ && $authors~/\,/ {
# extract words from author affiliation and compare with the countries.
# If a match is found increment that array entry.
w = split($authors, b, ",")
for (i=0;i<w;i++){
gsub(";"," ",b[i])
if (b[i] in a) a[b[i]]++
}
}
END {
for(k in a){
if(a[k]) print a[k], k
}
}
# HOW TO RUN:
# awk -v topic=birds -f prob3.awk cities.txt \
../mag_papers_allcols/mag_papers_*.allcols.txt \
../aminer_papers_allcols_excl/aminer_papers_*.allcols.excl.txt
# awk -v topic=birds -f prob3.awk countries.txt ...
# awk -v topic=birds -f prob3.awk universities.txt ...
# Run the following pipeline on the results:
# sort -nr -k 1 citywise_papers.txt > tmp && mv tmp citywise_papers.txt
# OR
# After running the swift app:
# awk -F: '{a[$2]+=$1} END {for (k in a) print a[k],k}' joined_cities.txt \
| sort -nr > tmp && mv tmp joined_cities.txt
Problem 4
Identify how topics have shifted over time.
This problem may be solved in three distinct ways. The first approach processes the database to find out year-wise occurrence of any given two topics together. It generates a list of years and the number of times both topics has occurred in a single publication in that year. For example, the plot shown below shows how the terms “obesity” and “sugar” have trended together in publications over the years.
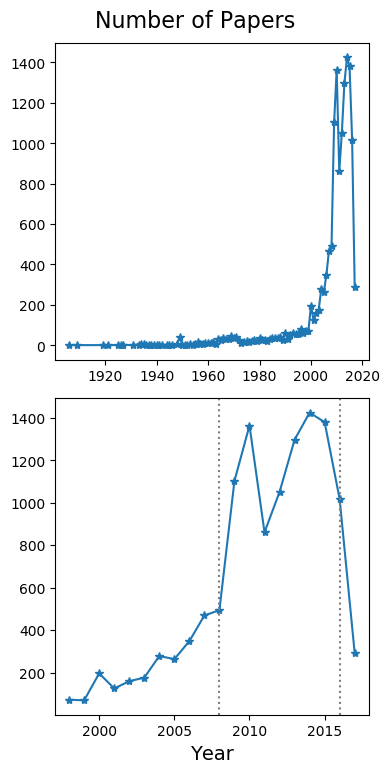
Awk code shown below.
#!/usr/bin/env awk -f
# Problem Statement
# Identify how topics have shifted over time.
# Solution 1 below will search for any two topics
# mentioned and show the number of occurrence of both the topics year-wise
BEGIN {
# Field Separator
FS = "qwqw"
IGNORECASE = 1
# Field names
id=1; title=2; num_authors=3; doi=4; fos_isbn=5; doctype_issn=6;
lang=7; n_citation=8; issue=9; url=10; volume=11; page_start=12;
page_end=13; year=14; venue=15; publisher_pdf=16; references=17;
keywords=18; abstract=19; authors=20;
}
$lang~/en/ && $year!~/null/ && $0~topic1 && $0~topic2 {
a[$year]++
}
END {
n = asorti(a,b)
printf("Trend for topics: %s, %s\n", topic1, topic2)
for (i=1;i<=n;i++) printf("%d :- %d\n", b[i], a[b[i]])
}
# Run as follows:
# awk -v topic1=obesity -v topic2=sugar -f code/prob4.awk aminer_mag_papers/*.txt
The second approach finds the papers that has highest impact in each year and
extracts the keywords in those papers. The impact is computed by the paper that
is cited the most in that year. The result for this task are in
results/yearwise_trending_keywords.txt in the form of year, keywords,
citations triplet.
#!/usr/bin/env awk -f
# Problem Statement
# Identify how topics have shifted over time.
# Solution 2 is to find the highest cited paper
# year-wise and figure out the topics it was based on
BEGIN {
FS = OFS = "qwqw"
IGNORECASE = 1
# Field names
id=1; title=2; num_authors=3; doi=4; fos_isbn=5; doctype_issn=6;
lang=7; n_citation=8; issue=9; url=10; volume=11; page_start=12;
page_end=13; year=14; venue=15; publisher_pdf=16; references=17;
keywords=18; abstract=19; authors=20;
}
$lang~/en/ && $year!~/null/ && $year<2020 && $keywords!~/null/
&& $n_citation!~/null/ && $n_citation>max[$year] {
max[$year] = $n_citation; a[$year]=$keywords
}
END {
n = asorti(a,b)
for (i=1;i<=n;i++) print b[i], a[b[i]], max[b[i]]
}
# Run via Swift in parallel. If serial, run like so:
# awk -f code/prob4_p2.awk aminer_mag_papers/*.txt > yearwise_trending_keywords.txt
The third approach finds the top 10 most frequently occurring terms each year
to find how the topics get in and out of trend over the years. An mkv animation
video showing a bubble plot of words trending between the year 1800 and 2017 is
here.
A file list of all the words is found in results/trending_words_by_year. A
snapshot trending words bubble in 2002 is shown below:
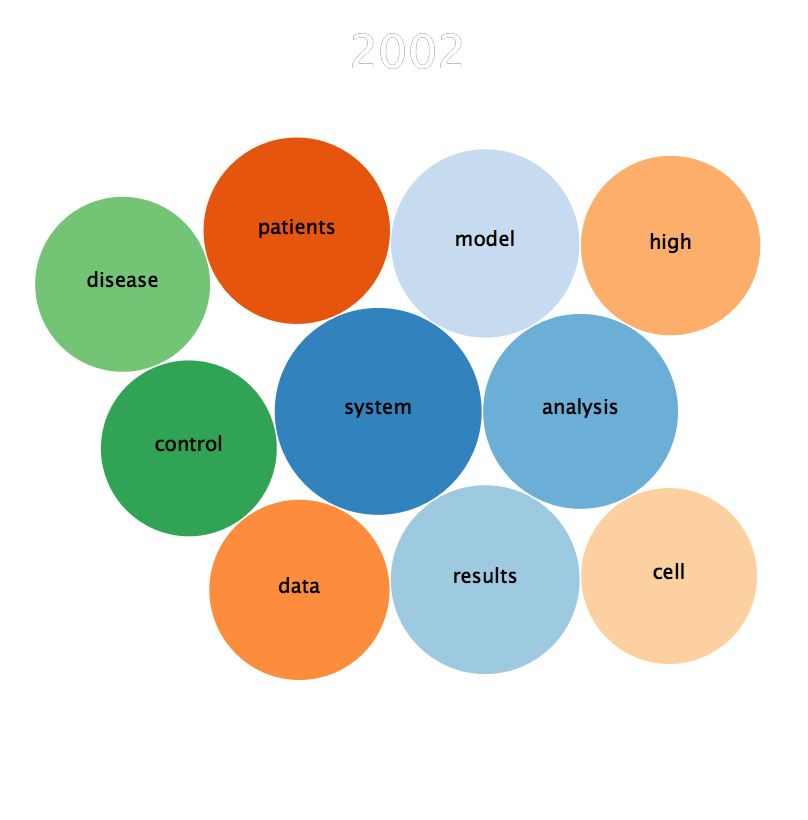
The Awk code that generates the raw data for above picture is shown below:
#!/usr/bin/env awk -f
# find the top 10 trending topics year-wise and see how they appear/disappear in the trend
# We achieve this by writing keywords, titles and abstract to files named after
# the year they appeared and do postprocessing on those files
BEGIN {
FS = "qwqw"
IGNORECASE = 1
# Field names
id=1; title=2; num_authors=3; doi=4; fos_isbn=5; doctype_issn=6;
lang=7; n_citation=8; issue=9; url=10; volume=11; page_start=12;
page_end=13; year=14; venue=15; publisher_pdf=16; references=17;
keywords=18; abstract=19; authors=20;
}
#collect stop words
NR == FNR {x[$1];next}
$lang~/en/ && $n_citation>0 && $year==yr && $keywords!~/null/{
# write title, keywords and abstract to a file
# titled by the year in which they appear
# treat title
$title = tolower($title)
split($title, a, " ")
for (i in a) if(length(a[i])>2 && match(a[i],/[a-z]/) && a[i] in x == 0) print a[i]
# treat keywords
$keywords = tolower($keywords)
gsub("\"","",$keywords)
split($keywords, b, ",")
for (i in b) if(length(b[i])>2 && match(b[i],/[a-z]/) && b[i] in x == 0) print b[i]
# treat abstract
$abstract = tolower($abstract)
gsub("\"","",$abstract)
gsub(","," ",$abstract)
split($abstract, c, " ")
for (i in c) if(length(c[i])>2 && match(c[i],/[a-z]/) && c[i] in x == 0) print c[i]
}
#Do the following for postprocessing:
#for i in 18?? 19?? 20??
# do (grep -o -E '\w+' $i | tr [A-Z] [a-z] \
# | sed -e 's/null//g' -e 's/^.$//g' -e 's/^..$//g' -e 's/^[0-9]*$//g' \
# | awk NF | fgrep -v -w -f stop_words.txt \
# | sort | uniq -c | sort -nr \
# | head -10 > trending/trending.$i.txt) & done
Parallelizing the third approach was challenging as it involved a two-level nested foreach loop. The outer loop iterates over the years and the inner loop iterates over the input files. The HPC implementation finishes in 48 minutes. Swift code for this shown below.
import files;
import io;
import unix;
import string;
app (file out) myawk (file awkprog, file infile, file stopwords, string yr){
"/usr/bin/awk" "-v" yr "-f" awkprog stopwords infile @stdout=out
}
file aminer[] = glob("/dev/shm/aminer_mag_papers/*.txt");
foreach y in [1800:2017:1]{
file yearfiles[];
foreach v, i in aminer{
yearfiles[i] = myawk(input("/home/km0/SMC18/src/prob4_p3.awk"),
v,
input("/home/km0/SMC18/data/stop_words.txt"),
sprintf("yr=%s",toString(y)));
}
file joined <sprintf("year%s.txt",toString(y))> = cat(yearfiles);
}
Problem 5
Given a research proposal, determine whether the proposed work has been accomplished previously.
This has a simple solution: Find the keywords on a new proposal. If those
keywords appear in an existing publication record, it is a suspect. A broad
list of suspects may be found with logical OR between keywords which could
be narrowed down with logical AND. The keywords may be arbitrarily combined
in ORs and ANDs. The results file /results/suspects.txt shows over 1,400
suspects for an AND combination of keywords: battery, electronics,
lithium, and energy from English language papers. The HPC implementation
finishes in 26 seconds. Awk code below.
#!/usr/bin/env awk -f
# Problem Statement
# Given a research proposal, determine whether the proposed work has been
# accomplished previously.
# Solution: Find the keywords in the new proposal.
# If those keywords appear in an existing publication record, it is a suspect.
BEGIN {
FS = "qwqw"
IGNORECASE = 1
# Field names
id=1; title=2; num_authors=3; doi=4; fos_isbn=5; doctype_issn=6;
lang=7; n_citation=8; issue=9; url=10; volume=11; page_start=12;
page_end=13; year=14; venue=15; publisher_pdf=16; references=17;
keywords=18; abstract=19; authors=20;
}
# topic1 .. topic4 are provided at command line
$0~topic1 && $0~topic2 && $0~topic3 && $0~topic4 && $lang~/en/ && $authors!~/null/{
print $id, $title, $authors, $year
}
Conclusion
I show how the classic Unix tools may be leveraged to solve modern problems and that millions of records may be processed in under a minute at scale. About the data itself, it seems the biosciences research dominates the publications followed by perhaps physics. I am sure more sophisticated tools could be used to get refined results and gain better insights – this is my take. I won the data challenge. Awk is awesome!